Papers
Transforming cabbage into turnip: a polynomial algorithm for sorting signed
S. Hannenhalli, P.A. Pevzner (1999), Journal of the ACM, 46, 1-27.
This paper introduced the first polynomial algorithm for analyzing genome rearrangements. This algorithm was applied to studies of rearrangements in cancer and developing genome rearrangement web servers <a href="http://grimm.ucsd.edu/cgi-bin/grimm.cgi">MGR</a> (Bourque and Pevzner, Genome Res., 2001) and <a href="http://mgra.cblab.org">MGRA</a> (Alekseyev and Pevzner, Genome Res., 2009).
An Eulerian path approach to DNA fragment assembly
P.A. Pevzner, H. Tang, M.S. Waterman (2001), Proceedings of the National Academy of Sciences, 98, 9748-9753.
The de Bruijn-based genome assembly approach outlined in this paper is now at the heart of modern sequencing algorithms; the lion's share of genomes assembled today use the algorithmic paradigm proposed in this paper.
Human and mouse genomic sequences reveal extensive breakpoint reuse in mam
P.A. Pevzner and G. Tesler (2003), Proceedings of the National Academy of Sciences. 100, 7672-7677.
This paper has refuted the Random Breakage Model of chromosome evolution and introduced the new Fragile Breakage Model. It is a rare example when an established biological theory was refuted using an algorithmic analysis. This work contributed to our studies of rearrangements in large genomics projects aimed at sequencing mouse (Waterston et al., Nature, 2002), rat (Gibbs et al., Nature 2004), and chicken (Hillier et al., Nature 2004) genomes.
SPAdes: a new genome assembly algorithms and its applications to single cel
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012), J Comput Biol., 19:455-77.
This paper introduced the SPAdes genome assembler, the most widely used assembler today with over 20,000 citations to date.
Assembly of long, error-prone reads using repeat graphs
M. Kolmogorov, J. Yuan, Y. Lin, and P.A. Pevzner (2019) Nature Biotechnology
This paper introduced the fast and accurate Flye long-read genome assembler that constructs the repeat graph of the genome and facilitates analysis of mosaic segmental duplication.
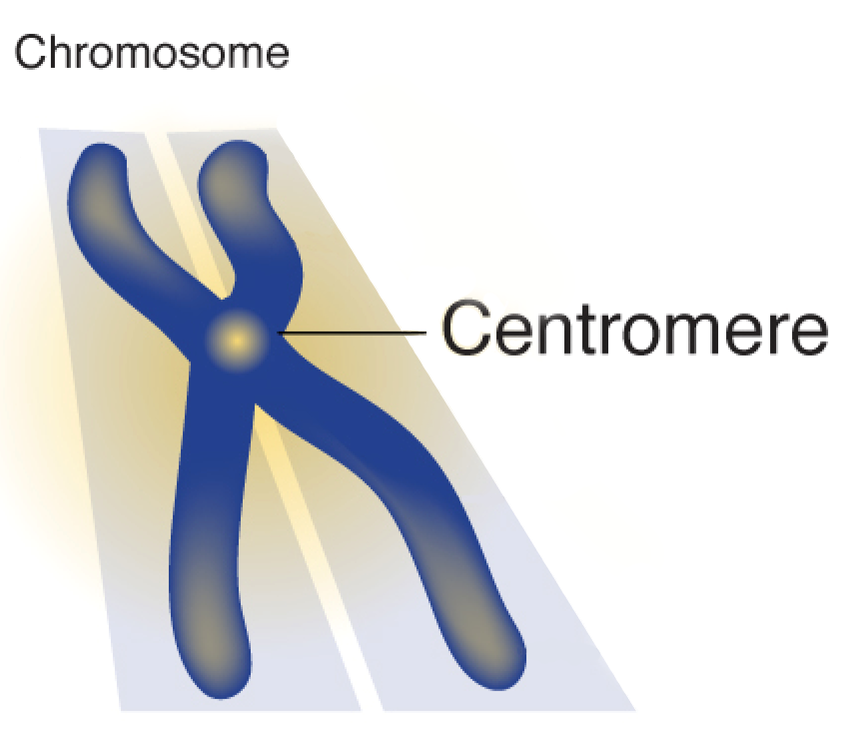
UniAligner: a parameter-free framework for fast sequence alignment
A. Bzikadze and P. A. Pevzner. UniAligner: a parameter-free framework for fast sequence alignment. Nature Methods 2023
Even though the recent advances in ‘complete genomics’ revealed the previously inaccessible genomic regions, analysis of variations in centromeres and other extra-long tandem repeats (ETRs) faces an algorithmic challenge since there are currently no tools for accurate sequence comparison of ETRs. Counterintuitively, the classical alignment approaches, such as the Smith–Waterman algorithm, fail to construct biologically adequate alignments of ETRs. This paper present UniAligner—the first sequence alignment algorithm that addresses the challenge of aligning highly-repetitive genomic regions.